An overview of processing free text in the electronic health record
Note: This blog post is an excerpt from the forthcoming book A Researcher's Guide to Using Electronic Health Records, From Planning to Presentation, 2nd edition.
Extracting meaning from free text is a challenging yet common problem in electronic health record (EHR) research. In fact, a whole sub-field of computer science is concerned with methods for addressing this problem, which range from string parsers and regular expressions to probabilistic techniques including natural language processing (NLP). The goal of parsing narrative free text in the EHR is to extract relevant information for epidemiologic analysis that may not be captured in discrete or coded fields. Textual data captures information such as patient-reported outcomes, symptoms, illness trajectory and progress notes, differential diagnoses, social and behavioral history, and so on. It is mostly non-redundant to other information in the EHR, and as such it has tremendous value for public health and epidemiologic measures not normally found in the EHR, such as the social determinants of health. However, there are numerous challenges to processing free text. These challenges include the breadth of medical terminology and abbreviations; misspellings, unconventional sentence structure, and phrases; documentation patterns specific to clinician, specialty, location, and EHR; and complexity and ability to validate more sophisticated approaches including NLP. This blog post serves as a high level overview of a very complex field.

String parsing
To begin we will consider the more straightforward approaches to manipulating text. These approaches assume that the free text has been extracted from the EHR and exist as a string-type variable in the research database or spreadsheet. There are a variety of functions that exist in statistical software to manipulate and parse strings, including retrieving the length of a string, sub-setting or splitting a string, concatenating multiple strings together, locating the position or matching one or more characters within a string, truncating or removing a portion of the string, replacing a sequence of characters in a string with another, and standardizing the case or removing special characters. There are many other string manipulation functions and their availability depends on the statistical software in use.
Let us consider an example of a hepatitis C test result received from an outside laboratory and stored as free text in the EHR: "HCV antibody POSITIVE; HCV RNA POSITIVE 900,000 IU/L; Interpretation: chronic infection with hepatitis C." There are several approaches we may take to extracting meaning from this string. On the one hand, we may be interested in a qualitative interpretation: the patient has a chronic hepatis C infection. On the other hand, we may be interested in a quantitative interpretation: the patient's viral load is high at 900,000 copies. For this hypothetical, we are interested in operationalizing two variables: hepatitis C antibody presence and, if positive, hepatitis C viral load. Permutations of these variables can be used to infer no infection, prior infection, or current infection, and thus these two variables are sufficient for the presumed analytic goals.
The text string includes delimiters - characters that separate data points, a semicolon in this case - and splitting the string by this delimiter will result in three new strings: "HCV antibody POSITIVE", "HCV RNA POSITIVE 900,000 IU/L", and "Interpretation: chronic infection with hepatitis C". To focus on the viral load, we now need to extract the actual value. One approach for this would be to use a string search function that identifies numbers in text (i.e., regular expression, covered later); a second option may be to take the "word" immediately preceding the unit of measure "IU/L"; and a third option may be to further parse by a space delimiter and extract the penultimate string. The validity of these approaches depends on the consistency of the lab reporting format. Following extraction of "900,000" we may need to use a character replacement function to remove the comma, otherwise the value may not be castable as a numeric data type. This can be further complicated by patients who do not have detectable virus in their blood where the lab report may say "HCV RNA NEGATIVE Undetectable". A frequency table can be used to identify all the possible permutations and code can be written to handle the special cases. As can be seen from this trivial example, string parsing is as much an art as it is a science, and what is easy for humans to accomplish through manual chart review can be quite challenging for a computer to automate. Fortunately, there are tools available to aid in the process.
For smaller research databases, Microsoft Excel offers many of the string manipulation functions previously discussed but in a graphical user interface obviating the need for complicated programming. For example, the Text to Columns function can accomplish the delimiter-based parsing needed to separate antibody and viral load from the other attributes in the test result. A visual inspection of the data will allow the researcher to align the parsed fields into the appropriate columns (variables) and search for inconsistencies. Then, using the Filter operation on the column, outlier or nonconforming values can be isolated and addressed. While Excel makes this process intuitive, it adds additional time to the data preprocessing and will be counterproductive on large datasets.
When the use of Excel is not an option, regular expressions are a powerful alternative for retrieving patterns in text, however constructing regular expressions requires experience with their syntax. If a recurring pattern exists in the free text, regular expression searches can parse the required data. Returning to the earlier example, "HCV antibody POSITIVE; HCV RNA POSITIVE 900,000 IU/L; Interpretation: chronic infection with hepatitis C.", the regular expression
.*[ANTIBODY|Antibody|antibody]\s(POSITIVE|NEGATIVE|positive|negative).*[RNA|Rna|rna]\s(POSITIVE|NEGATIVE|positive|negative)\s(\d+(,\d{3})*|UNDETECTABLE|Undetectable|undetectable)
will automatically parse 1) the results of the antibody test as positive or negative, 2) the RNA test as positive or negative, and 3) the viral load whether a number or undetectable. This regular expression allows variation in capitalization as well as the use of commas in the number and can be further enhanced to consider a multitude of reporting differences. Regular expression search engines are available in many statistical software packages
Natural language processing
The hepatitis C lab result example in the previous section was simplistic and not representative of the reality of free text responses. Suppose we needed to operationalize a variable for male patients that captures whether they have sex with other men. Assuming sexual orientation or behavior is not captured in the EHR as a discrete variable, this information is most likely found as a free text note in the patients' social histories. Examples of such language may include: "Pt reports hx of sex with men," "Pt is MSM", "Pt identifies as homosexual", "Pt has same gender partner". In these cases, a standard text parser would be difficult to implement. We may develop a list of key words to search for in the free text note, such as "MSM", "gay", or "homosexual," and while this may work for positive documentation, or documentation that occurs in the presence of a finding, this would fail to detect the subtleties of negative documentation, or documentation that occurs in the absence of a finding: "Pt denies same sex behavior." Further, this information may be buried in other notes taken during the patient history requiring more sophisticated processing techniques such as NLP.
There are multiple types and sub-fields of NLP, including classification and extraction. Herein we focus on high level applications applied to the clinical narrative. NLP can also range from more simplistic rule-based approaches to ML algorithms, with hybrid adaptions of both. Several NLP solutions have been developed explicitly for the EHR. These EHR-specific solutions include home grown and open-source solutions such as the clinical text analysis knowledge extraction system (cTAKES) and the clinical event recognizer (CLEVER), as well as commercial options such as Linguamatics and Spark NLP for Healthcare. There are additionally a variety of open-source NLP libraries available for proficient programmers. For a historical review of NLP solutions in healthcare, refer to Savova et al.. Use of NLP tools depend on the free text being available and electronically extractable from the EHR; in the case of older paper-based records (that may or may not have been scanned into the EHR), optical character recognition will need to be applied first to digitize their contents prior to NLP.
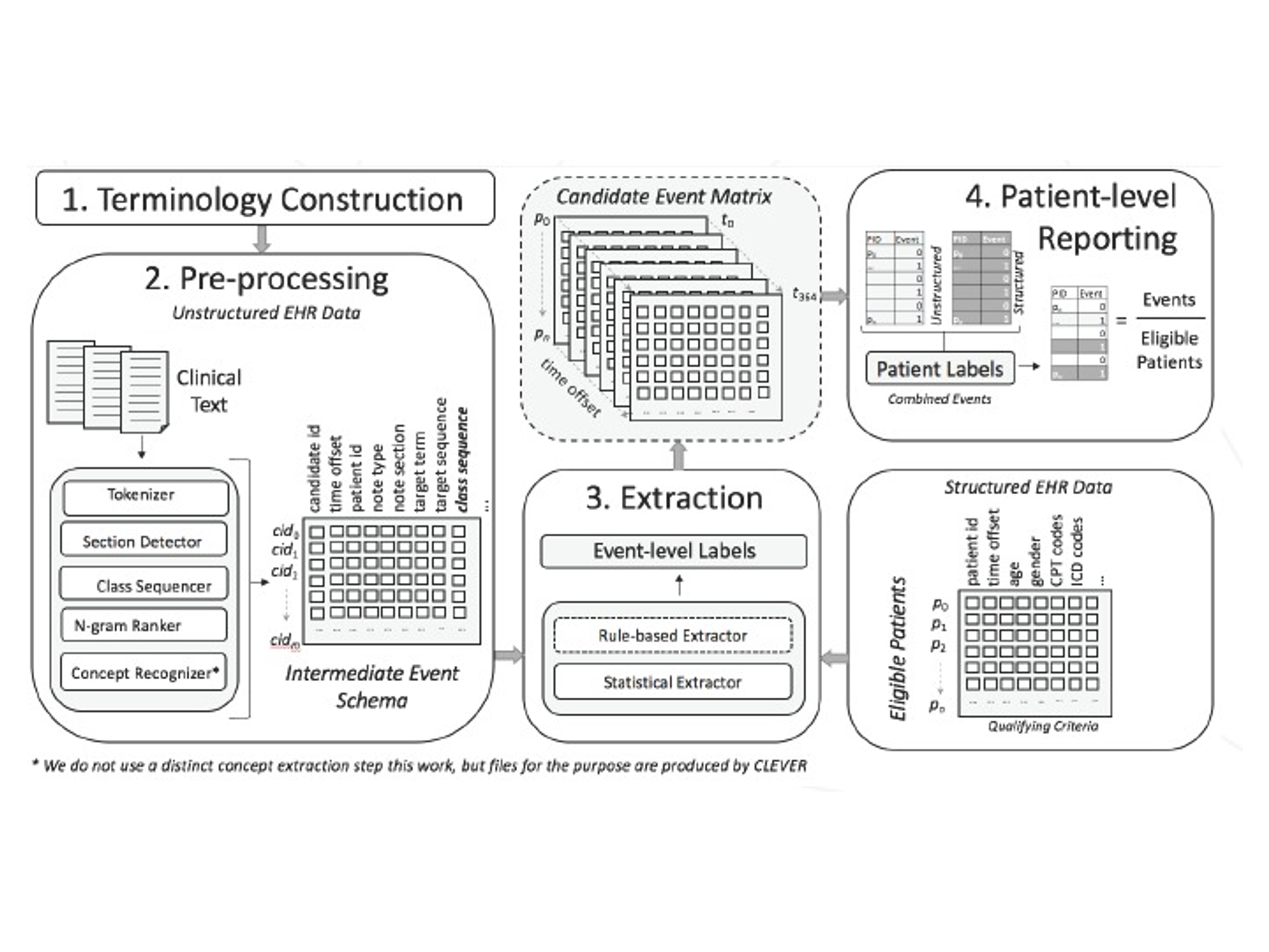
cTAKES is a hybrid rule-based and ML NLP approach for processing clinical narratives (Savova et al.). It processes text in a sequential and cumulative fashion (figure below). At a high level, the narrative is parsed into sentences; words, numbers, and dates are identified and normalized to standardize tense, spelling, and so on; terminology is mapped into the National Library of Medicine's Unified Medical Language System (UMLS); and annotations are applied to understand context, such as the negative or positive occurrence of a condition. In several validation studies, compared to an expert-abstracted gold standard measure, cTAKES achieved agreement better than 90% for ascertaining cardiovascular risk factors for a case-control study of peripheral arterial disease and treatment classification for a pharmacogenomics breast cancer treatment study (Savova et al.). The cTAKES software is now part of an open source consortium and regularly updated.

CLEVER is a tool that aids in the annotation of EHR narratives for clinical concepts. The first step in the CLEVER pipeline (figure below) is to create the dictionary of terms applicable to the extraction based on expert opinion. For example, for researchers studying lupus in the EHR, this dictionary would include terms indicative of the condition. As with cTAKES, this seed dictionary is referenced against the UMLS to build a list of synonymous terms. The clinical narrative should first be cleaned for punctuation and normalized, and then the CLEVER tool can be applied to generate the annotations. Annotations will flag positive mentions of disease, for example "patient has lupus", or negative mentions of disease, for example "rule out lupus". Although CLEVER is still being used for NLP due to its efficiency and simplicity, it is not being actively maintained as of writing. Other NLP annotation engines have been deployed into EHRs, such as the UMLS MetaMap. Researchers have compared the accuracy in the extraction of clinical notes of these open-source NLP engines.
There are abundant examples in the literature of NLP applied to free text in the EHR, many of which have been summarized in systematic reviews (Shivade et al., 2014; Ford et al., 2016; Kreimeyer et al., 2017; Wang et al., 2018; Koleck et al., 2019; Sheikhalishahi et al., 2019; Spasic & Nenadic, 2020). While there are domain specific reviews of NLP (Abbe et al., 2016; Spasic et al., 2014; Pons et al., 2016; Juhn & Liu, 2020), herein we focus on more general applications of NLP for clinical narratives in the English language. Koleck et al. (2019) reviewed the use of NLP to extract symptoms captured in free text, such as "pain, fatigue, disturbed sleep, depressed mood, anxiety, nausea, dyspnea, and pruritus". Given their subjective nature that differs by provider and clinical specialty, symptoms could be documented in varied ways, thus hampering more straightforward data mining techniques. Among the 27 reviewed relevant articles that met the inclusion criteria, the authors observed a wide variety of NLP approaches used by the researchers including commercial software, open-source algorithms, and, in approximately 50% of the studies, homegrown solutions. Manual chart review served as the gold standard for comparison in most studies and the authors noted that most of the reviewed studies' objectives were to identify symptomology in the EHR, which in their view, suggested that NLP was a nascent field concerned with methodology over application.
Sheikhalishahi et al. (2019) conducted a review on the application of NLP to identify chronic disease, because in their experience, unstructured data was far more common than structured data for documentation of diseases such as heart disease, stroke, cancer, diabetes, and lung disease. Among 106 reviewed articles covering 43 diseases in 10 disease categories, the authors found that metabolic diseases tended to have the greatest amount of structured data while diseases of the circulatory system had the greatest amount of unstructured data. As with Kolek et al., Sheikhalishahi et al. observed a diversity of NLP algorithms, with an increasing trend towards ML - in particular, support vector machines and naive Bayes - or hybrid approaches in more recent years. Nevertheless, rule-based approaches were still quite common likely due to their relative simplicity despite the improved accuracy of what are known as deep classifiers or general deep learning methods for NLP. The authors also noted that use of NLP for identifying co-morbidities or combining structured and unstructured data for prediction and longitudinal modeling was underutilized. In addition to calling for improvement in temporal extraction, they also called for focusing NLP to describe relationships between conditions and diagnoses beyond phenotype identification, and more public unstructured data for algorithmic development.
Spasic and Nenadic (2020) conducted a review of 110 articles focused on ML NLP in clinical narratives. Despite rich datasets being available in the reviewed studies, supervised ML was often used on subsets of the available data, owing to the burden of creating a validation dataset via manual chart review. This was identified as a major bottleneck to further development of ML for NLP. In general, the data requirements for supervised learning were more likely to be met for in-hospital death, discharge, readmission, and emergency department visits. An additional finding was the common use of single institution EHRs, which may limit the transferability of the ML approach given how clinical narratives, among other factors, vary by institution (Sohn et al., 2018). Figure 4 in Spasic and Nenadic summarizes the clinical applications of NLP in the reviewed studies: the most common applications were enabling other tasks (i.e., NLP serving as a steppingstone, often to diagnosis), prognosis, clinical phenotyping, and care improvement. These applications most commonly mined EHR data in clinical notes, admission notes, discharge summaries, progress reports, radiology reports, allergy entries, and free text medication orders.
To conclude this post, we will focus on two studies that have mined EHR data for social determinants that are otherwise infrequently captured via diagnostic or billing codes: homelessness (Bejan et al., 2018) and social isolation (Zhu et al., 2019). In the study by Bejan et al., the authors sought to identify homelessness from over 100 million clinical notes for 2.6 million patients in the Vanderbilt University Medical Center EHR, while comparing two data driven NLP methodologies: lexical associations, a measure of connection between words and phrases, and word2vec, an open-source ML NLP toolkit. Seed terms were initially provided by the researchers, and the NLP engines expanded the query to form the search lexicon. The clinical notes were then compared against the vocabulary, results weighted and filtered to remove negative documentation, and subsequently assessed for relevance. Expert opinion was used to refine terms and manual review compared NLP annotations with manual annotation, resulting in the final search lexicon: homeless, homelessness, shelter, unemployed, jobless, and incarceration. Performance of the ML NLP approach surpassed the lexical association approach, with area under the curves exceeding 0.90. Based on their success, the authors further explored identifying childhood adverse experiences from the EHR and suggested that other social of determinants can be operationalized from unstructured EHR data using NLP.
Indeed, this was the case with Zhu et al. who sought to identify another social determinant - social isolation - in the clinical notes of patients with prostate cancer at the Medical University of South Carolina. As opposed to Bejan et al., where the authors used homegrown or open-source NLP, Zhu et al. relied on the commercial NLP software Linguamatics (IQVIA, Durham, NC). Their approach followed the standard NLP pipeline: social isolation seed terms were provided based on context area expertise and then expanded in the NLP software, with subsequent expert review. Once a final lexicon was developed, the algorithm was deployed on a training set and performance tested against a testing data set, both of which included the gold standard manual chart review annotations. Continual refinement occurred to maximize the accuracy of the algorithm. In total, 200,000 clinical notes for 4,000 patients were mined for social isolation. These notes came from multiple places in the EHR: progress notes, telephone encounters, plan of care documents, consults, history and physical notes, discharge summaries, and emergency department notes. The sensitivity of their algorithm was 97% and the positive predictive value was 90%, leading the authors to conclude that when social isolation information is documented in the EHR, NLP is a viable option for extraction.
Despite the success of these two studies, researchers should keep in mind that 1) these are single center studies that 2) take resources to create the gold standard and expertise to implement NLP, and 3) the context or reasons for the social determinants will not be automatically parsed from the free text. Further, 4) the tuning of these algorithms must balance potential false positives and false negatives on a case-by-case basis, and 5) NLP applications are still largely in the exploratory and algorithmic development stages.
Cite: Goldstein ND. An overview of processing free text in the electronic health record. Nov 8, 2022. DOI: 10.17918/goldsteinepi.